Difference between revisions of "Training exercises"
| (20 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
== FoldX energies == | == FoldX energies == | ||
| + | Before we start, some basic information about FoldX energies is necessary. | ||
| + | |||
| + | First of all, FoldX energies are expressed in kcal/mol. | ||
| + | |||
The main focus of FoldX is the prediction of free energy '''changes''', e.g. what happens to the free energy of the protein when we mutate an Asp to a Tyr? FoldX will then calculate the free energy of the wild type (WT) and the mutant (MT) and make the difference: | The main focus of FoldX is the prediction of free energy '''changes''', e.g. what happens to the free energy of the protein when we mutate an Asp to a Tyr? FoldX will then calculate the free energy of the wild type (WT) and the mutant (MT) and make the difference: | ||
| − | <math>\Delta\Delta</math>G(change) = <math>\Delta</math>G( | + | <div style="background:#FFDDFF;border:1px solid #FFC0CB;"> |
| + | <math>\Delta\Delta</math>G(change) = <math>\Delta</math>G(MT) - <math>\Delta</math>G(WT) | ||
| + | </div> | ||
FoldX is trained to make <math>\Delta\Delta</math>G(change) approach experimental values. It is important to realize that <math>\Delta</math>G(WT) and <math>\Delta</math>G(MT) are meaningless numbers as such. These do not correlate with experimental values. Only <math>\Delta\Delta</math>G(change) does. | FoldX is trained to make <math>\Delta\Delta</math>G(change) approach experimental values. It is important to realize that <math>\Delta</math>G(WT) and <math>\Delta</math>G(MT) are meaningless numbers as such. These do not correlate with experimental values. Only <math>\Delta\Delta</math>G(change) does. | ||
| Line 11: | Line 17: | ||
As a rule of thumb we use: | As a rule of thumb we use: | ||
| + | <div style="background:#FFDDFF;border:1px solid #FFC0CB;"> | ||
<math>\Delta\Delta</math>G(change) > 0 : the mutation is destabilizing | <math>\Delta\Delta</math>G(change) > 0 : the mutation is destabilizing | ||
<math>\Delta\Delta</math>G(change) < 0 : the mutation is stabilizing | <math>\Delta\Delta</math>G(change) < 0 : the mutation is stabilizing | ||
| + | </div> | ||
The error margin of FoldX is approximately 0.5 kcal/mol, so changes in that range are insignificant. | The error margin of FoldX is approximately 0.5 kcal/mol, so changes in that range are insignificant. | ||
== Minimizing the structure == | == Minimizing the structure == | ||
| − | FoldX assumes that the starting structure has been energy minimized. Although crystal structures with high resolution represent | + | FoldX assumes that the starting structure has been energy minimized. Although crystal structures with high resolution often represent a low energy form, FoldX performs best when we minimize it just before we do the predictions. This FoldX procedure is called '''RepairPDB''' and should be done on each structure you want to perform calculations on. |
Download and open the YASARA scene [[Media:2AC0.sce|2AC0.sce]] in YASARA. This is a part of a tetrameric complex of the transcription factor P53 bound to DNA. I removed 3 of the 4 P53 structures for simplicity and visualized some nice features. | Download and open the YASARA scene [[Media:2AC0.sce|2AC0.sce]] in YASARA. This is a part of a tetrameric complex of the transcription factor P53 bound to DNA. I removed 3 of the 4 P53 structures for simplicity and visualized some nice features. | ||
| Line 27: | Line 35: | ||
File > Load > YASARA Scene | File > Load > YASARA Scene | ||
</pre> | </pre> | ||
| + | |||
| + | [[File:training_1.png|none|thumb|500px|P53 monomer bound to DNA]] | ||
To Repair (or minimize) the structure with FoldX go to: | To Repair (or minimize) the structure with FoldX go to: | ||
| Line 34: | Line 44: | ||
</pre> | </pre> | ||
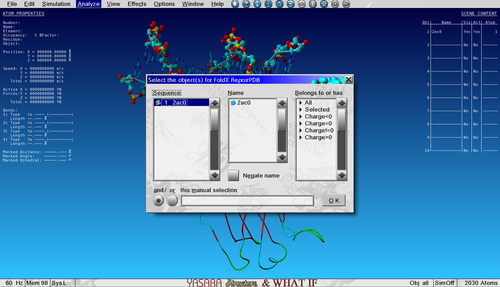
| − | And select the only object in the list. | + | [[File:training_2.png|none|thumb|500px|Select the object for repairing]] |
| + | |||
| + | And select the only object in the list. Repairing a structure can take a few minutes depending on the size of the protein. | ||
When the Repair is finished, the Repaired Object is placed in Object 2 (see top right corner) and superposed with the original Object 1. Take a look at the sidechains and see what FoldX has done while Repairing. | When the Repair is finished, the Repaired Object is placed in Object 2 (see top right corner) and superposed with the original Object 1. Take a look at the sidechains and see what FoldX has done while Repairing. | ||
| Line 44: | Line 56: | ||
Because we will continue working with this Repaired Object, we can now hide the entire Object 1 by toggling the '''Vis'''ibility column in the top right corner head-up display (HUD). | Because we will continue working with this Repaired Object, we can now hide the entire Object 1 by toggling the '''Vis'''ibility column in the top right corner head-up display (HUD). | ||
| − | == | + | '''As repairing can take quite some time, it is a good idea to save each repaired structure for re-use later on. Save the repaired structure as a PDB file with:''' |
| + | |||
| + | <pre class="ex">File > Save as > PDB file</pre> | ||
| + | |||
| + | and select the second Object in the leftmost Sequence column. | ||
| + | |||
| + | == Predict effect of a point mutation on protein structure stability == | ||
Turn on all the atoms in the DNA chain. These are chains/molecules G and H. | Turn on all the atoms in the DNA chain. These are chains/molecules G and H. | ||
<pre class="ex"> | <pre class="ex"> | ||
| Line 50: | Line 68: | ||
</pre> | </pre> | ||
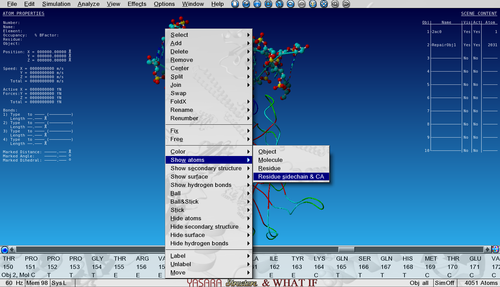
| − | Show the sidechain of Ala159 in the core of P53 by searching for it in the sequence selector (in Object 2!), right-click on it and go to: | + | Show the sidechain of Ala159 in the core of P53 by searching for it in the sequence selector (in Object 2, not Object 1!), right-click on it and go to: |
<pre class="ex"> | <pre class="ex"> | ||
Show atoms > Residue sidechain and CA | Show atoms > Residue sidechain and CA | ||
</pre> | </pre> | ||
| + | |||
| + | [[File:training_3.png|none|thumb|500px|Hovering the mouse at the bottom of the screen will open the sequence selector. Search for Ala 159 in Object 2, not Object 1!]] | ||
| + | [[File:training_4.png|none|thumb|500px|Right-clicking on Ala159 opens the residue-context menu]] | ||
<pre class="ex"> | <pre class="ex"> | ||
| Line 66: | Line 87: | ||
FoldX > Mutate residue | FoldX > Mutate residue | ||
</pre> | </pre> | ||
| + | |||
| + | [[File:training_5.png|none|thumb|500px|Right-click on Ala159 and select "FoldX > Mutate residue" to mutate this specific residue]] | ||
A number of menus is now presented and here is what you need to do in each menu: | A number of menus is now presented and here is what you need to do in each menu: | ||
| Line 73: | Line 96: | ||
# 'Move neighbours' and 'Show VdW clashes in WT and mutant' | # 'Move neighbours' and 'Show VdW clashes in WT and mutant' | ||
# Don't change any numerical options in the last menu | # Don't change any numerical options in the last menu | ||
| + | |||
| + | [[File:training_6.png|none|thumb|500px|View of the first options menu with Move neighbours and VdW clashes selected]] | ||
| + | |||
| + | More information on all these selection menus is explained in detail in the [[FoldX_tools_in_YASARA#Mutate_residue | mutate residue section of the guide]]. | ||
== Analyze the mutation == | == Analyze the mutation == | ||
FoldX has mutated the Ala to Trp and the structure with the Trp mutation has been loaded in the next Object (3) and is superposed with the wild type (WT, Object 2). We selected an option to show the VdW clashes in WT and mutant. The atoms that give rise to steric clashes are colored in <span style="color:red">'''red'''</span>. Toggle the '''Vis'''ibility of Object 2 (WT) and Object 3 (mutant) and see how many clashes we introduced by mutating the Ala to Trp.<br> | FoldX has mutated the Ala to Trp and the structure with the Trp mutation has been loaded in the next Object (3) and is superposed with the wild type (WT, Object 2). We selected an option to show the VdW clashes in WT and mutant. The atoms that give rise to steric clashes are colored in <span style="color:red">'''red'''</span>. Toggle the '''Vis'''ibility of Object 2 (WT) and Object 3 (mutant) and see how many clashes we introduced by mutating the Ala to Trp.<br> | ||
| + | |||
| + | [[File:training_7.png|none|thumb|500px|Zoomed-in-view on the original Ala159, no Vander Waals clashes here]] | ||
| + | [[File:training_8.png|none|thumb|500px|Zoomed-in-view on the mutated Ala159Trp, lots of red Vander Waals clashes here]] | ||
<pre class="ex"> | <pre class="ex"> | ||
| Line 88: | Line 118: | ||
</pre> | </pre> | ||
| − | == | + | [[File:training_9.png|none|thumb|500px|In the console (to open press spacebar twice), we see an energy change of +29 kcal/mol. Clearly, this is severely destabilising the structure]] |
| + | |||
| + | == Predict effect of a point mutation on protein-DNA binding == | ||
Hide Object 3 by toggling its '''Vis'''ibility so that only Object 2 (the repaired WT) is visible.<br> | Hide Object 3 by toggling its '''Vis'''ibility so that only Object 2 (the repaired WT) is visible.<br> | ||
First turn on all atoms in the molecules G and H (DNA) again as you did previously, because the FoldX run has hidden it (it rearranged the view to show the VdW clashes). | First turn on all atoms in the molecules G and H (DNA) again as you did previously, because the FoldX run has hidden it (it rearranged the view to show the VdW clashes). | ||
| Line 103: | Line 135: | ||
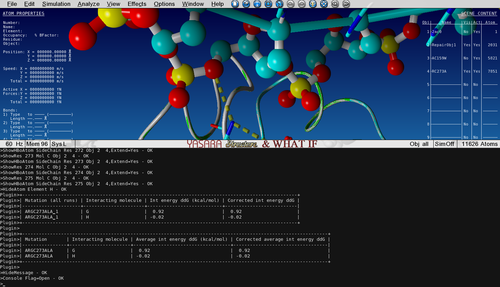
'''Notice how the positively charged Arginine is making an electrostatic interaction with the negative phosphate from the DNA backbone.''' | '''Notice how the positively charged Arginine is making an electrostatic interaction with the negative phosphate from the DNA backbone.''' | ||
| + | |||
| + | [[File:training_10.png|none|thumb|500px|R273 makes an electrostatic interaction with the DNA phosphate groups.]] | ||
Let's see what would happen to the interaction energy between the DNA and P53 when we mutate this Arginine to Alanine. | Let's see what would happen to the interaction energy between the DNA and P53 when we mutate this Arginine to Alanine. | ||
| Line 118: | Line 152: | ||
# 'Move neighbours' and 'Show disrupted and new hydrogen bonds' | # 'Move neighbours' and 'Show disrupted and new hydrogen bonds' | ||
# Don't change any numerical options in the last menu | # Don't change any numerical options in the last menu | ||
| + | |||
| + | [[File:training_11.png|none|thumb|500px|View of the first options menu with "Show new and disrupted hydrogen bonds" selected.]] | ||
Toggle the '''Vis'''ibility between this mutant and the WT structure and see how the hydrogen bonding changes. | Toggle the '''Vis'''ibility between this mutant and the WT structure and see how the hydrogen bonding changes. | ||
| Line 124: | Line 160: | ||
Check the output in the Console to see what the change in interaction energy is between P53 and DNA chain G upon mutation. | Check the output in the Console to see what the change in interaction energy is between P53 and DNA chain G upon mutation. | ||
</pre> | </pre> | ||
| + | |||
| + | [[File:training_12.png|none|thumb|500px|We see that the mutation decreases the interaction with DNA strand G by approximately 1 kcal/mol (we lost 1 hydrogen bond).]] | ||
<pre class="ex"> | <pre class="ex"> | ||
| Line 130: | Line 168: | ||
<pre>Instead of DNA-protein, FoldX can of course also calculate interaction energy changes in protein-protein or peptide-protein complexes.</pre> | <pre>Instead of DNA-protein, FoldX can of course also calculate interaction energy changes in protein-protein or peptide-protein complexes.</pre> | ||
| + | |||
| + | == Running FoldX from the command line == | ||
| + | In case you need to run many calculations, it can be handy to know how to run FoldX from the command line and steer it with a script for batch runs. <br> | ||
| + | You need four files to do a command line run: | ||
| + | |||
| + | # a PDB file | ||
| + | # a FoldX commands file | ||
| + | # a FoldX options file | ||
| + | # rotabase.txt file (comes with FoldX) | ||
| + | |||
| + | Getting the PDB file is up to you. For the other files, you can re-use commands and option files generated by the FoldX plugin for YASARA. If you need to do a RepairPDB e.g., just do such a run with the plugin first. Then go to: | ||
| + | <pre> | ||
| + | Analyse > FoldX > Save last calculation | ||
| + | </pre> | ||
| + | |||
| + | This saves all output files including the commands and options files. Go to the folder where you just saved everything and look for the ''commands_repair.txt'' and ''options_repair.txt'' files. Make sure you have the above 4 files in the same folder. Then you can fire the (example) command: | ||
| + | |||
| + | <pre> | ||
| + | foldx -manual Object1.pdb options_repair.txt commands_repair.txt | ||
| + | </pre> | ||
| + | |||
| + | This should give you the same results as you would run it with the plugin. | ||
| + | |||
| + | <pre> | ||
| + | The options used by the plugin are only a basic (but solid) set of configurable options. See the FoldX manual for all options. | ||
| + | </pre> | ||
Latest revision as of 08:39, 22 April 2011
Contents
Introduction
In this section we will let the FoldX plugin loose on some real world examples and give you step-by-step instructions on how to proceed and analyze the results. We will use the P53 tumor suppressor protein as our example molecule. In a first exercise we will show you how to make a point mutation with FoldX and how to determine if the mutation is stabilizing or destabilizing for the P53 structure. In a second exercise we will design a mutation in the P53 structure at the DNA binding interface and determine how the mutation affects the interaction energy of P53 with the DNA strand.
FoldX energies
Before we start, some basic information about FoldX energies is necessary.
First of all, FoldX energies are expressed in kcal/mol.
The main focus of FoldX is the prediction of free energy changes, e.g. what happens to the free energy of the protein when we mutate an Asp to a Tyr? FoldX will then calculate the free energy of the wild type (WT) and the mutant (MT) and make the difference:
<math>\Delta\Delta</math>G(change) = <math>\Delta</math>G(MT) - <math>\Delta</math>G(WT)
FoldX is trained to make <math>\Delta\Delta</math>G(change) approach experimental values. It is important to realize that <math>\Delta</math>G(WT) and <math>\Delta</math>G(MT) are meaningless numbers as such. These do not correlate with experimental values. Only <math>\Delta\Delta</math>G(change) does.
As a rule of thumb we use:
<math>\Delta\Delta</math>G(change) > 0 : the mutation is destabilizing
<math>\Delta\Delta</math>G(change) < 0 : the mutation is stabilizing
The error margin of FoldX is approximately 0.5 kcal/mol, so changes in that range are insignificant.
Minimizing the structure
FoldX assumes that the starting structure has been energy minimized. Although crystal structures with high resolution often represent a low energy form, FoldX performs best when we minimize it just before we do the predictions. This FoldX procedure is called RepairPDB and should be done on each structure you want to perform calculations on.
Download and open the YASARA scene 2AC0.sce in YASARA. This is a part of a tetrameric complex of the transcription factor P53 bound to DNA. I removed 3 of the 4 P53 structures for simplicity and visualized some nice features.
Load the scene with:
File > Load > YASARA Scene
To Repair (or minimize) the structure with FoldX go to:
Analyze > FoldX > Repair object
And select the only object in the list. Repairing a structure can take a few minutes depending on the size of the protein.
When the Repair is finished, the Repaired Object is placed in Object 2 (see top right corner) and superposed with the original Object 1. Take a look at the sidechains and see what FoldX has done while Repairing.
If you feel the repair takes too long (more than 10 minutes) due to a slow computer, download and open this YASARA Scene with the Repaired Object.
File > Load > YASARA Scene
Because we will continue working with this Repaired Object, we can now hide the entire Object 1 by toggling the Visibility column in the top right corner head-up display (HUD).
As repairing can take quite some time, it is a good idea to save each repaired structure for re-use later on. Save the repaired structure as a PDB file with:
File > Save as > PDB file
and select the second Object in the leftmost Sequence column.
Predict effect of a point mutation on protein structure stability
Turn on all the atoms in the DNA chain. These are chains/molecules G and H.
View > Show atoms in > Molecule > Name > G and H
Show the sidechain of Ala159 in the core of P53 by searching for it in the sequence selector (in Object 2, not Object 1!), right-click on it and go to:
Show atoms > Residue sidechain and CA
Zoom in on the Ala159 by keeping the CTRL button pressed and at the same time left click on it in the sequence selector.
We are going to mutate this residue to the largest natural amino acid, tryptophan (W).
Right-click again on Ala159 in Object 2 in the sequence selector and go to:
FoldX > Mutate residue
A number of menus is now presented and here is what you need to do in each menu:
- Only select Calculate stability change
- Select Trp
- 'Move neighbours' and 'Show VdW clashes in WT and mutant'
- Don't change any numerical options in the last menu
More information on all these selection menus is explained in detail in the mutate residue section of the guide.
Analyze the mutation
FoldX has mutated the Ala to Trp and the structure with the Trp mutation has been loaded in the next Object (3) and is superposed with the wild type (WT, Object 2). We selected an option to show the VdW clashes in WT and mutant. The atoms that give rise to steric clashes are colored in red. Toggle the Visibility of Object 2 (WT) and Object 3 (mutant) and see how many clashes we introduced by mutating the Ala to Trp.
Van der Waals clashes are red colored atoms. Do you see a difference around the mutation site between WT and mutant? Toggle the Visibility of WT and mutant to see the differences.
Open the Console by pressing the spacebar twice and see the free energy change of the mutation. Anything above a change of +0.5kcal/mol is already assumed to be destabilizing.
Is the Ala195Trp mutation destabilizing or stabilizing?
Predict effect of a point mutation on protein-DNA binding
Hide Object 3 by toggling its Visibility so that only Object 2 (the repaired WT) is visible.
First turn on all atoms in the molecules G and H (DNA) again as you did previously, because the FoldX run has hidden it (it rearranged the view to show the VdW clashes).
Show the sidechain of Arg273 of Object 2 by searching for it in the sequence selector, then right-click on it and go to:
Show atoms > Sidechain and CA
Zoom in on Arg273
Notice how the positively charged Arginine is making an electrostatic interaction with the negative phosphate from the DNA backbone.
Let's see what would happen to the interaction energy between the DNA and P53 when we mutate this Arginine to Alanine.
Right-click on this Arg273 in the sequence selector and go to:
FoldX > Mutate residue
A number of menus is now presented and here is what you need to do in each menu:
- Select Calculate interaction energy change
- Select Ala
- 'Move neighbours' and 'Show disrupted and new hydrogen bonds'
- Don't change any numerical options in the last menu
Toggle the Visibility between this mutant and the WT structure and see how the hydrogen bonding changes.
Check the output in the Console to see what the change in interaction energy is between P53 and DNA chain G upon mutation.
Why doesn't the mutation affect the interaction with DNA chain H?
Instead of DNA-protein, FoldX can of course also calculate interaction energy changes in protein-protein or peptide-protein complexes.
Running FoldX from the command line
In case you need to run many calculations, it can be handy to know how to run FoldX from the command line and steer it with a script for batch runs.
You need four files to do a command line run:
- a PDB file
- a FoldX commands file
- a FoldX options file
- rotabase.txt file (comes with FoldX)
Getting the PDB file is up to you. For the other files, you can re-use commands and option files generated by the FoldX plugin for YASARA. If you need to do a RepairPDB e.g., just do such a run with the plugin first. Then go to:
Analyse > FoldX > Save last calculation
This saves all output files including the commands and options files. Go to the folder where you just saved everything and look for the commands_repair.txt and options_repair.txt files. Make sure you have the above 4 files in the same folder. Then you can fire the (example) command:
foldx -manual Object1.pdb options_repair.txt commands_repair.txt
This should give you the same results as you would run it with the plugin.
The options used by the plugin are only a basic (but solid) set of configurable options. See the FoldX manual for all options.